install.packages("cleanslate")15 Running environment
In the previous chapter, we generated instructions to manually create the running environments for reproducing the A&R deliverables.
In this chapter, we focus on automating the creation of the R environments with R code to accelerate the dry run testing process, simplifying the ADRG instructions, and making it easy to recreate different environment settings with reproducible analysis results.
15.1 Prerequisites
cleanslate is an R package that offers a solution to create portable R environments.
As of Q4 2021, the cleanslate package used in this chapter is still under active development and validation. This chapter gives a preview of the planned APIs. They may change in the future.
Install cleanslate from CRAN (once available):
Or from GitHub (once available):
remotes::install_github("Merck/cleanslate")15.2 Practical considerations
The cleanslate package supports:
- Creating a project folder with project-specific context (
.Rproj,.Rprofile,.Renviron) - Installing a specific version of R into the project folder
- Installing a specific version of Rtools into the project folder
An essential feature of cleanslate is that it does not require administrator privileges to run R and Rtools installers. This makes it easier to deploy under enterprise settings and avoids security and portability concerns.
As many of the A&R deliverables are currently created, validated, and delivered under Windows, the primary focus is Windows at the moment, while the support for other platforms might be added in future versions.
15.3 Create canonical environments
One can create a running environment with “canonical” settings with a single function call to use_cleanslate():
cleanslate::use_cleanslate(
"C:/temp/",
r_version = "4.1.1",
from = "https://cran.r-project.org/",
repo = "https://packagemanager.posit.co/cran/2021-08-06"
)This will
- Create a project folder under
C:/temp/with a.Rprojfile; - Download R
4.1.1installer from CRAN, and install it intoC:/temp/R/; - Not install Rtools (by default,
rtools_version = NULL); - Create a
.Rprofilefile under the project folder, setoptions(repos)to use the specifiedrepo(a Posit Public Package Manager snapshot in this example), and give instruction to set the R binary path in RStudio IDE; - Create a
.Renvironfile under the project folder and set the library path to be the library of the project-specific R installation.
As a principle, one should always double-click the .Rproj file to open the project. This will ensure some sanity checks in the .Rprofile, such as whether the R and library are located within the project folder.
15.4 Create tailored environments
To create a more customized running environment, one can use the specific functions to tailor each aspect, for example:
library("cleanslate")
"C:/temp/" %>%
use_project() %>%
use_rprofile() %>%
use_renviron() %>%
use_r_version(version = "4.1.1") %>%
use_rtools(version = "rtools40")The project context functions (use_project(), use_rprofile(), use_renviron) support custom templates using brew.
The use_r_*() functions have variations that serve as shortcuts to use R versions defined by release lifecycles, for example, use_r_release(), use_r_oldrel(), and use_r_devel(). Note that to ensure better reproducibility, one should still use use_r_version() as the release, oldrel, and devel versions will shift as time goes by.
The helper functions version_*() and snapshot_*() can assist you in determining specific versions of R and Rtools that are currently available, besides generating and verifying the snapshot repo links.
15.5 Update ADRG
If you use cleanslate, remember to update the ADRG instructions for executing the analysis programs in R. Mostly, this can simplify the first three steps on creating a project, installing a specific version of R, and configuring the package repo location. For example:
Appendix: Instructions to Execute Analysis Program in R
1. Setup R environment
Open the existing R, install the required packages by running the code below.
install.packages("cleanslate")
Create a temporary working directory, for example, "C:\tempwork".
Copy all submitted R programs into the temporary folder.
In the same R session, run the code below to create a project
with a portable R environment.
cleanslate::use_cleanslate(
"C:/temp/",
r_version = "4.1.1",
from = "https://cran.r-project.org/",
repo = "https://packagemanager.posit.co/cran/2021-08-06"
)
2. Open the project
Go to the working directory created above, double click the .Rproj file
to open the project in RStudio IDE. Follow the instructions to select the
project-specific R version, then restart RStudio IDE. If successful,
the R version and package repo should be printed as defined above.
3. Install open-source R packages
In the new R session, install the required packages by running the code below.
install.packages(c("pkglite", "publicpkg1", "publicpkg2"))
4. Install proprietary R packages
All internal R packages are packed in the file r0pkgs.txt. In the same R session,
restore the package structures and install them by running the code below.
Adjust the output path as needed to use a writable local directory.
pkglite::unpack("r0pkgs.txt", output = ".", install = TRUE)
5. Update path to dataset and TLFs
INPUT path: to rerun the analysis programs, define the path variable
- Path for ADaM data: path$adam
OUTPUT path: to save the analysis results, define the path variable
- Path for output TLFs: path$output
All these paths need to be defined before executing the analysis program. For example:
path = list(adam = "/path/to/esub/analysis/adam/datasets/") # Modify to use actual location
path$outtable = path$outgraph = "." # Outputs saved to the current folder
6. Execute analysis program
To reproduce the analysis results, rerun the following programs:
- tlf-01-disposition.txt
- tlf-02-population.txt
- tlf-03-baseline.txt
- tlf-04-efficacy.txt
- tlf-05-ae-summary.txt
- tlf-06-ae-spec.txt15.6 RStudio addin
To make it convenient to use cleanslate in experiments, one can also use its RStudio IDE addin. After cleanslate is installed, click Addins -> cleanslate -> Create portable R environment in RStudio IDE, or call cleanslate:::create_env_addin() to open it.
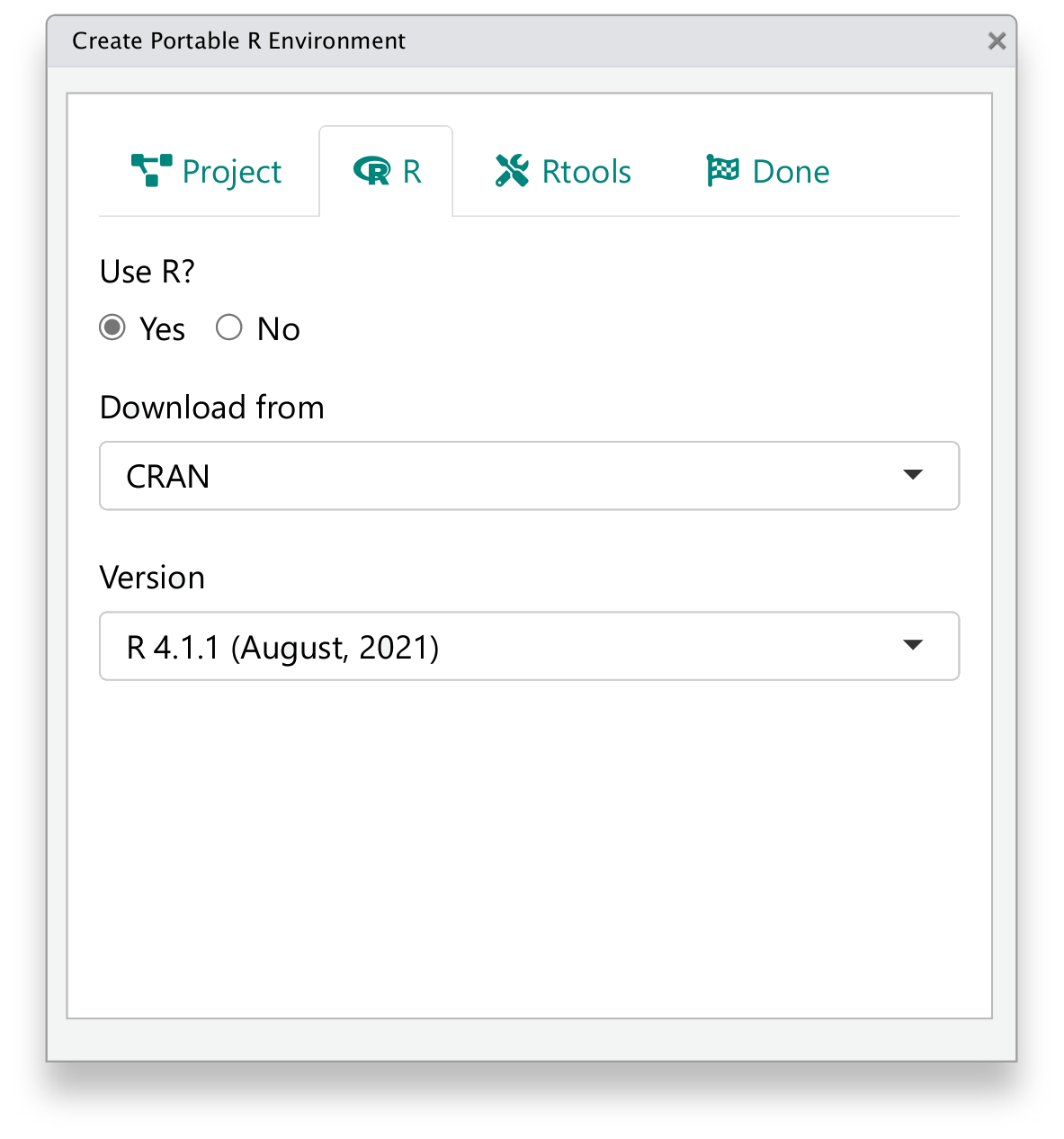
The addin provides a wizard-like interface to help create the environment with the most important options, yet with less flexibility compared to the functional API demonstrated above.